Inside AlphaFold: DeepMind’s Recipe For Success
How To Build Teams Destined For Breakthroughs
Get a list of personally curated and freely accessible ML, NLP, and computer vision resources for FREE on newsletter sign-up.
Consider sharing this with someone who wants to know more about machine learning.
At a major computer vision conference, I had the privilege to attend a keynote from Pushmeet Kohli (and it was very kind of him to interact later in person and respond to questions from a few of us after the keynote) one of the people behind DeepMind’s AlphaFold and VP of research at Google DeepMind.
Of course, the technical details that went into solving the problem are important and I will leave those for another time. In this piece, I want to go over what it takes to set up a team to take on this massive interdisciplinary challenge and come away with success.
I would like to share with you what I learned from that very valuable interaction with Pushmeet.
0. Introduction
AlphaFold, developed by DeepMind, has revolutionized the field of protein structure prediction. By predicting the 3D structures of proteins from their amino acid sequences, AlphaFold addresses a longstanding challenge in biology.
This breakthrough has profound implications for biomedical research, disease understanding (like understanding protein structure during the COVID-19 pandemic), and biotechnology. Beyond its technical achievements, the AlphaFold project offers invaluable lessons on problem-solving, team management, and interdisciplinary collaboration.
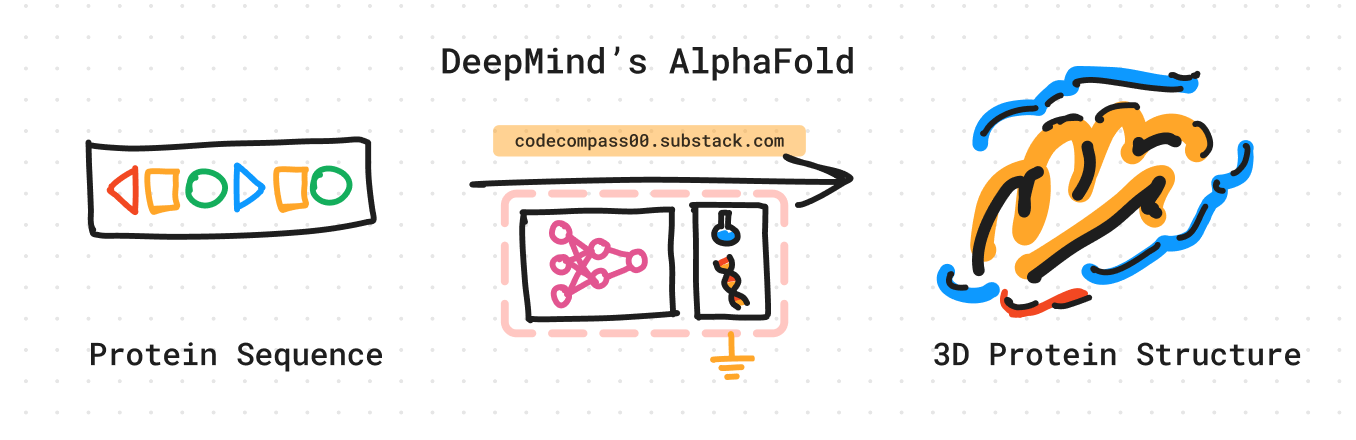
1. What does AlphaFold do?
AlphaFold takes the amino acid sequence of a protein as its primary input and outputs a predicted 3D structure of that protein.
Input: An amino acid sequence of the protein of interest.
Output: Predicted 3D structure of the protein complex with atomic coordinates.
2. Relevance of Protein Structure Prediction
Proteins are essential molecules that perform various functions within living organisms. Their function is determined by their 3D structure, which is shaped by the sequence of amino acids they are composed of. Knowing the structure of a protein can take many months but when completed the structure provides insights into how it works and what it does. Accurate protein structure prediction is crucial because it can:
Accelerate Drug Discovery: By understanding protein structures, researchers can design more effective drugs.
Enhance Disease Understanding: Knowledge of protein structures can provide insights into the mechanisms of diseases, including COVID-19.
Advance Biotechnology: It allows for the design of novel enzymes and other proteins with specific functions.
In previous, iterations of the benchmark in protein folding, the CASP competition, there was a stable plateauing of the winning solutions around ~40.0. With DeepMind’s solution, it broke this barrier and jumped ahead of previous numbers by a significant margin. This newly set record was broken again with the second iteration: AlphaFold2.
Challenges and DeepMind’s Approach
The traditional methods of determining protein structures, such as X-ray crystallography and cryo-electron microscopy, are time-consuming and expensive. AlphaFold offers a scalable and efficient alternative, but developing such a sophisticated model came with its own set of challenges.
With LLMs on the rise, there is a need for proper benchmarks so you and I can know which model is better: OpenAI’s ChatGPT4, Google’s Gemini, or the new kid on the block from Meta’s LLAMA series.
Or do you want to read something more technical? Then read about Attention from the Transformers paper and how it changed machine learning forever. Yes, LLMs, I am looking at you!





!["Attention, Please!": A Visual Guide To The Attention Mechanism [Transformers Series]](https://substackcdn.com/image/fetch/$s_!mFbk!,w_140,h_140,c_fill,f_auto,q_auto:good,fl_progressive:steep,g_auto/https%3A%2F%2Fsubstack-post-media.s3.amazonaws.com%2Fpublic%2Fimages%2F375dc525-fa19-4e6f-81f2-68820bfd36a1_1903x856.png)